| |
Ph.D. Research Projects: |
| |
Picornaviruses and Their Receptor |

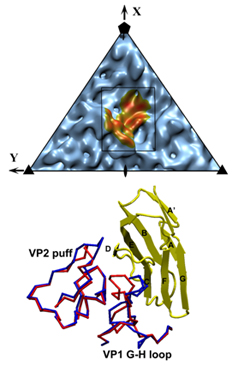 |
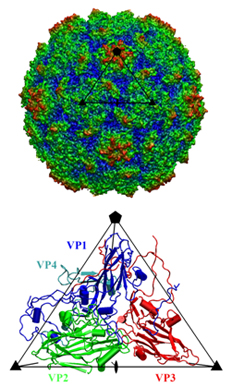
Coxackievirus A21 (CVA21) and human rhinoviruses (HRVs) are causative pathogens of the common cold. In the family of Picornaviridae, CVA21 and polioviruses belong to the Enterovirus genus, whereas rhinoviruses comprise a distinct genus. Nevertheless, CVA21 and major group HRV serotypes including 14 and 16 recognize intercellular adhesion molecule 1 (ICAM 1) as their cellular receptor, whereas polioviruses recognize poliovirus receptor (PVR). ICAM-1 is a cell surface glycoprotein that provides adhesion between leukocytes and endothelial cells. A variant of ICAM-1, ICAM-1Kilifi, is unable to form a stable complex with HRV16 but is still capable of binding CVA21 and HRV14. X-ray crystallography and cryo-electron microscope (cryo-EM) image analysis were combined to study the complex structures between ICAM-1 and each of the three viruses, CVA21, HRV14 and HRV16. The crystal structure of CVA21 was determined to 3.2 Å resolution. Three-dimensional structures of CVA21 complexed with ICAM 1Kilifi, of HRV14 and HRV16 each complexed with ICAM 1, and of HRV14 complexed with ICAM 1Kilifi, were determined by cryo-EM image reconstruction to between 8 and 13 Å resolution. The cryo-EM maps were fitted with the crystal structures of ICAM 1 and the corresponding viruses.The binding site of ICAM-1 to CVA21 or HRVs, and that of PVR to polioviruses overlap at the canyon region. Interactions within this common region may be essential for triggering viral destabilization after attachment to susceptible cells. However, the orientation of the receptor molecule relative to the virus surface is different for each virus. Significant differences in the structure of CVA21 with respect to the polioviruses account for the inability of ICAM 1 to bind polioviruses. Many virus/receptor contact residues are conserved within CVAs or HRVs but not between CVAs and HRVs, indicating that these two groups of viruses evolved separately to exploit different recognition modes of ICAM-1. Nevertheless, interfaces between the viruses and ICAM-1 display considerable shape and electrostatic complementarity and contain extensive ionic networks. The interactions between the viruses and ICAM 1Kilifi include one less salt bridge than with ICAM-1. As HRV16 has fewer overall interactions with ICAM-1 than HRV14, the absence of this salt bridge will have a greater impact on the binding of ICAM–1Kilifi to HRV16 than to HRV14. |
| |
Postdoctoral Research Project: |
| |
Structural Studies of the Giant Mimivirus |


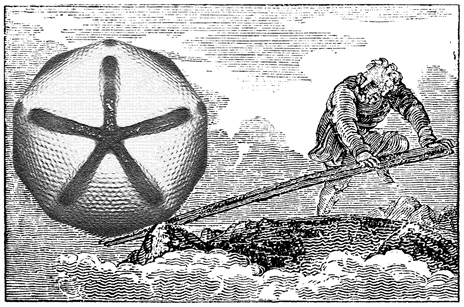 |
Mimivirus is the largest known virus whose genome and physical size are comparable to some small bacteria, blurring the boundary between a virus and a cell. Structural studies of Mimivirus have been difficult because of its size and long surface fibers. Here we report the use of enzymatic digestions to remove the surface fibers of Mimivirus, in order to expose the surface of the viral capsid. Cryo-electron microscopy (cryoEM) and atomic force microscopy were able to show that the 20 icosahedral faces of Mimivirus capsids have hexagonal arrays of depressions. Each depression is surrounded by six trimeric capsomers that are similar in structure to those in many other large, icosahedral double-stranded DNA viruses. Whereas in most viruses, these capsomers are hexagonally close-packed with the same orientation in each face, in Mimivirus there are vacancies at the systematic depressions with neighboring capsomers differing in orientation by 60°. The previously observed starfish-shaped feature is well-resolved and found to be on each virus particle and is associated with a special pentameric vertex. The arms of the starfish fit into the gaps between the five faces surrounding the unique vertex, acting as a seal. Furthermore, the enveloped nucleocapsid is accurately positioned and oriented within the capsid with a concave surface facing the unique vertex. Thus, the starfish-shaped feature and the organization of the nucleocapsid might regulate the delivery of the genome to the host. The structure of Mimivirus, as well as the various fiber components observed in the virus, suggests that the Mimivirus genome includes genes derived from both eukaryotic and prokaryotic organisms. |
| |
Current research projects: |
| |
Structural Studies of the Giant Marine Virus CroV |
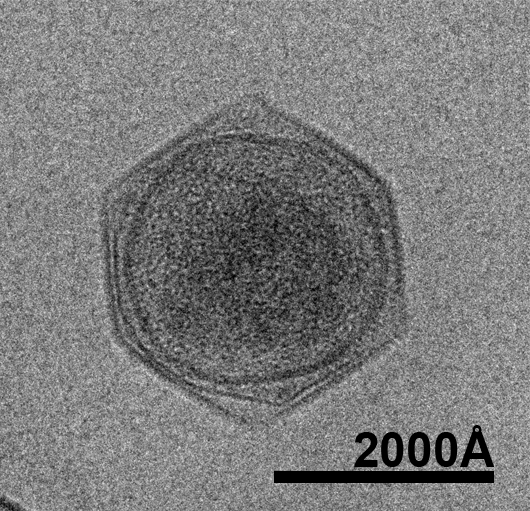 |
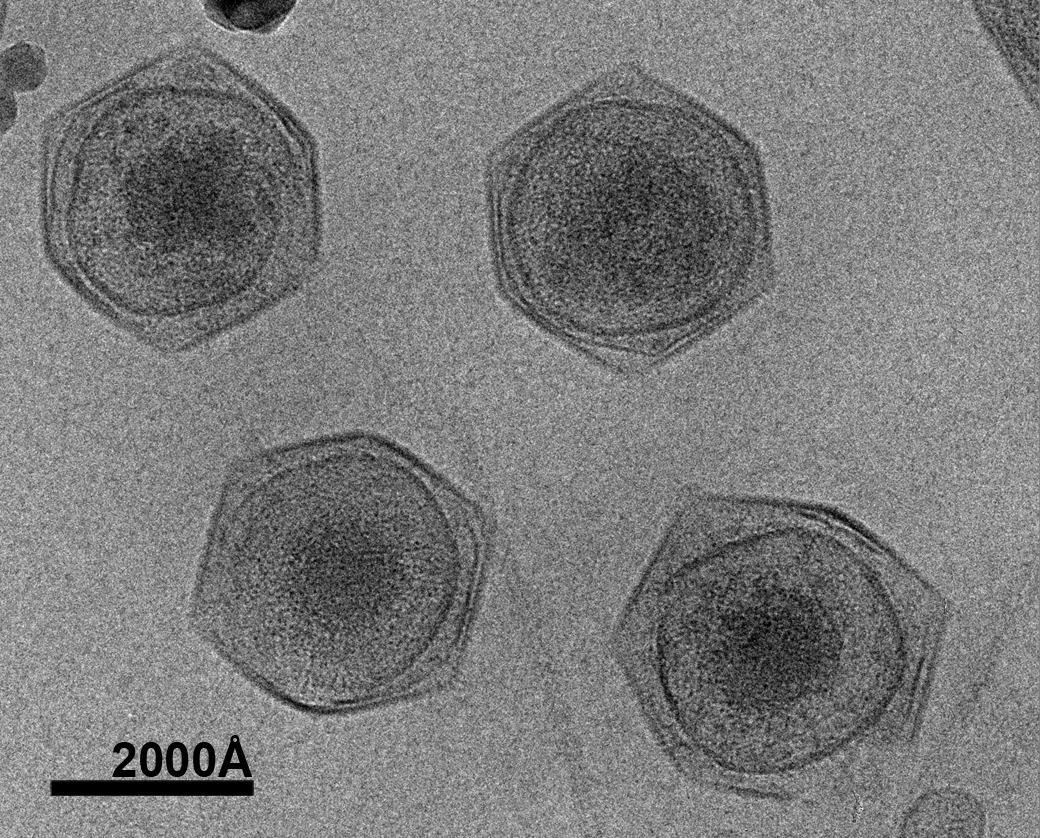
The oceans cover 70% of the earth’s surface. They not only play a key role in the global climate and ecological system but also supply human with biological and non-biological resources. Marine microorganisms produce 90% of the biomass in the ocean and about half of the earth’s oxygen. Only recently have scientists begun to realize the abundance of viruses in the ocean and their contribution to the marine nutrient cycles. The estimated 1030 total number of viruses in the oceans by far exceeds the total number of prokaryotes and protists combined. With about 1023 viral infections happening every second, viruses kill approximately 20% of the biomass generated by microorganisms every day. This has made marine virus one of the major driving forces in marine nutrient cycles. Marine viruses also sustain the biodiversity of the ocean through the mechanism of “killing the winner”, which prevents the dominance of a single marine species. Despite their important role in the ocean ecosystem, the majority of marine viruses have not been studied. Dr. Xiao decided to develop his future career in this unexplored area to determine the structures of marine viruses as a way to begin to understand their function and impact on oceanic ecosystems and biodiversity, which are linked to the overall health of the oceans.
The research goal is to determine the three-dimensional (3-D) pseudo-atomic structure of the intact CroV virus by combining cryo-electron microscopy (cryo-EM) and X-ray crystallography. In addition, the research project will initiate structural proteomics studies of CroV by first targeting important proteins of the virus. The elucidation of ultrastructural features of marine viruses will enhance our understanding of their life cycle and their interaction with their hosts, providing insight into their impact on the marine ecosystem. CroV, the largest marine virus known to date, has been chosen as the first representative model system not only because of its size and complexity but also because, by killing a major group of predators in marine microorganism communities, CroV has a significant impact on the oceans’ ecosystem and biodiversity. Functional predictions and analyses of the ultrastructural features of CroV will facilitate our understanding of its life cycle, its interaction with the hosts, and its roles in the ocean.
|
| |
Structural Studies of the Circadian Rhythm Related Proteins |

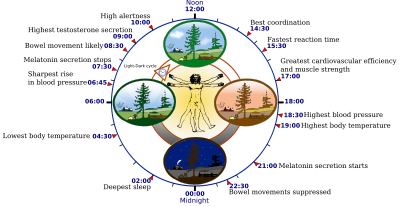 |
Circadian rhythm is an intrinsic roughly-24-hour biological clock embedded within most living organisms. In mammals, circadian rhythm coordinates sleep-wake cycles, blood pressure, body temperature and liver metabolism in a daily cycle. In humans, long term disruption of circadian rhythm can impair physical and mental health. For instance, sleep disorders are circadian related and affect about 20% of Americans, resulting in higher healthcare costs and lost productivity. Developing new therapeutic agents against circadian related diseases requires a better understanding of the mechanism involved in regulating the circadian rhythm. However, the lack of structural studies of the key functional components of the circadian rhythm limits our understanding of the molecular mechanism of this important clockwork. In order to overcome this barrier, combinations of structural biochemical tools, traditional biochemical, and biophysical methods will be applied to investigate the interactions among core regulatory components in the circadian clockwork. Conclusions regarding regulatory mechanisms of the circadian rhythm can be drawn from the various structures obtained, which will provide essential knowledge for future development of new therapeutic strategies against circadian disorders. |
| |
Structural Studies of the Assembly Process of CVA21 |
 |
In order to study virus assembly and infection, picornavirus was chosen as the research model because it is among the smallest and simplest viruses that includes a large knowledge base that has been accumulated from decades of research. coxsackievirus A21 (CVA21), which is one of the human common cold pathogen, has been chosen as the first target virus. CVA21 is closely related to polioviruses with about 80% genomic sequence identity and above 70% amino acid sequence identity in structural protein. For non-structural proteins, such as protease 3C and polymerase 3D, CVA21 has more than 90% amino acid identity to polioviruses. Notwithstanding the high sequence similarity between polioviruses and CVA21, polioviruses use the poliovirus receptor (PVR, CD155) instead of intercellular adhesion molecule 1(ICAM-1) or decay accelerating factor (DAF) as their cellular receptor. Transgenic mice expressing human ICAM-1 develop classic paralytic poliomyelitis after being infected with CVA21, indicating that CVA21 may share poliomyelitis potential and further supports the close relationship between poliovirus and CVA21. CVA21 was proposed as a replacement for poliovirus as the viral research system after the global poliovirus eradication. Binding to ICAM-1 is sufficient for CVA21 infection. However, binding of DAF does not trigger a conformational change of the CVA21 particles, nor is it sufficient to produce an infection, still requiring ICAM-1 for the cell entry. The major role of DAF may be to capture and concentrate infectious virions on the cell membrane. CVA21 is perhaps the only known virus that can bind to both DAF and ICAM 1. Because both these cell surface molecules are over-expressed on malignant melanoma cells, CVA21 has been used to selectively lyse tumor cells both in vitro and in vivo. Understanding the infection and assembly of CVA21 may also reveal its potential as a cancer treatment agent.
|
| |
Future Research Directions: |
| |
Structural Studies of the Plant Proteins |
 |
Dr. Xiao's research career started when he joined the Rice Genome Project Statellite Lab at Fudan University, Shanghai. There, he sequnced many Rice ETS and finished the first cDNA and genomic sequences of several Rice genes for the first time. Example includes Rice Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), a unique Calmodulin with myristoylation tail and the C-domain of V-ATPase. It was during the homology structural modeling research that made him change his career to determine the structure instead of to predict the structure. Hence, he pursued his Ph.D. in structural biochemistry. After obtaining the skills of structural dtermination techniques, Dr. Xiao wants to go back to study the structures of plant proteins that can improve our understanding of them and might help solving issues such as the global warming and global starvation.
|



























